U-Net: The Doctor’s New Digital Sidekick for Medical Imaging
Can AI help doctors see what even trained eyes might miss? Dive into how the Eff-UNet model pushes boundaries in pneumonia detection, bringing us closer to an AI-assisted future in diagnostics.

Medical image analysis, through modalities such as X-rays and MRI scans, remains crucial for the early detection of pathological anomalies. However, each scan requires careful human examination to identify the precise location and nature of irregularities, leading to significant time and resource overhead. With the advent of Artificial Intelligence, could this process be further streamlined? Convolutional Neural Networks (CNNs), a sophisticated extension of Artificial Neural Networks (ANNs), have demonstrated remarkable versatility across tasks such as image and signal classification. This article explores the potential of integrating these technologies into diagnostic workflows, aiming to enhance the speed and accuracy of patient diagnosis.
U-Net is a convolutional neural network (CNN) architecture, initially introduced by Ronneberger et al. in 2015, specifically designed for the task of image segmentation. Its distinct architecture is characterized by a symmetrical structure, comprising a contracting path that captures contextual information and an expansive path that facilitates precise localization. A key feature of U-Net is the inclusion of skip connections, which directly link feature maps from the contracting layers to their corresponding expanding layers. These connections play a critical role in improving segmentation accuracy by preserving fine-grained details across different spatial resolutions.
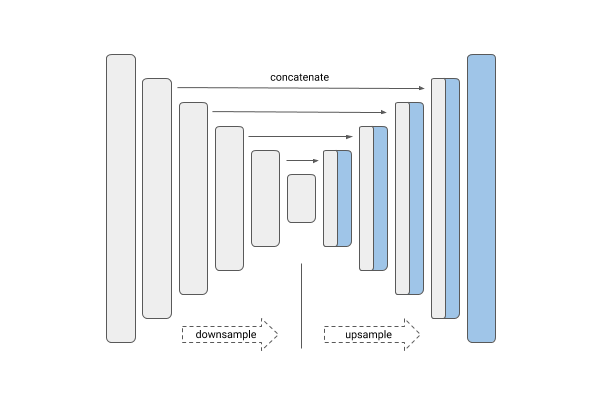
The design of U-Net enables it to effectively extract features from input images, often in conjunction with an input mask, through a contracting (downsampling) path. This is followed by an expansive (upsampling) path that reconstructs the image, restoring spatial resolution while preserving critical features.
Since its inception, numerous variations of the U-Net architecture have been proposed to further enhance its performance. Abd-Ellah et al. introduced the Two Parallel Cascaded U-Nets with Asymmetrical Residuals (TPCUAR-NET), which demonstrated high accuracy in the detection, classification, and segmentation of brain tumors. Similarly, Imtiaz et al. customized a U-Net model, achieving remarkable accuracy despite being trained on a relatively small dataset of 322 brain tumor MRI images.
Hybrid U-Net models, such as Eff-UNet introduced by Baheti et al., integrate EfficientNet for more efficient downsampling of input data. This architecture has been applied to various medical segmentation tasks, including pneumonia detection (Yu, 2021) and brain tumor segmentation (Lin & Lin, 2024), showcasing its versatility and effectiveness across diverse clinical applications.
These examples illustrate the potential of deep learning technologies to streamline and optimize processes in medical imaging tasks. The U-Net architecture, in particular, has demonstrated its effectiveness not only in full-image classification but also in more complex segmentation tasks. In the subsequent sections, this article will present a case study on image segmentation for pneumonia detection, utilizing the Eff-UNet architecture implemented with TensorFlow. Visual representations of the model’s learning progression during training will also be provided to offer deeper insights into its performance.
Methodology
To explore the capabilities of U-Net, specifically the Eff-UNet variant, we have chosen to utilize the segmentation_models library, focusing on their U-Net++ implementation. This model is executed using TensorFlow on the Google Colab platform. The primary objectives of this project are twofold: 1) to classify medical images and 2) to perform segmentation that reveals the "important features" identified by the model within the images. For this investigation, we have selected a lung X-ray dataset from Kaggle, which comprises X-ray images depicting both normal cases and instances of pneumonia for classification tasks. In the following section, I will outline the methodology employed in this experimental study.
Data
No Artificial Intelligence (AI) or Machine Learning (ML) model can be trained without the availability of data; thus, AI and ML serve as intelligent methodologies for conducting data analytics. To begin, we will undertake an Exploratory Data Analysis (EDA) of the dataset.
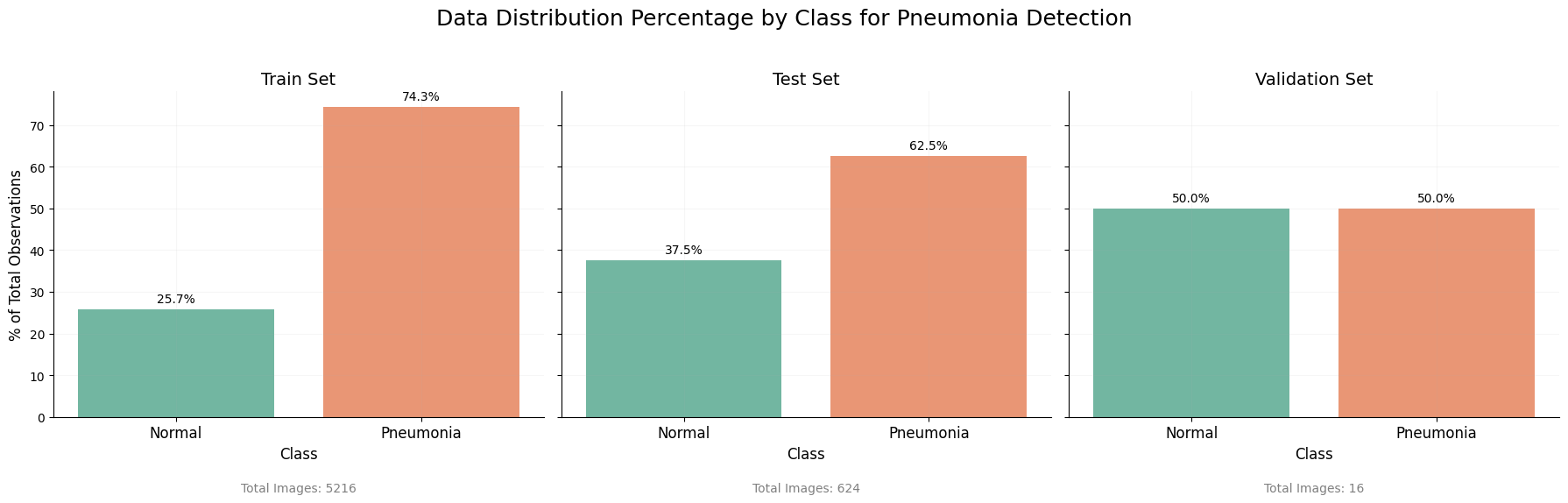
The distribution of classes within both the training and test sets is unequal, which may present challenges during the model training process. To maintain authenticity and reflect real-world scenarios, we will refrain from generating additional normal class images to achieve balance.
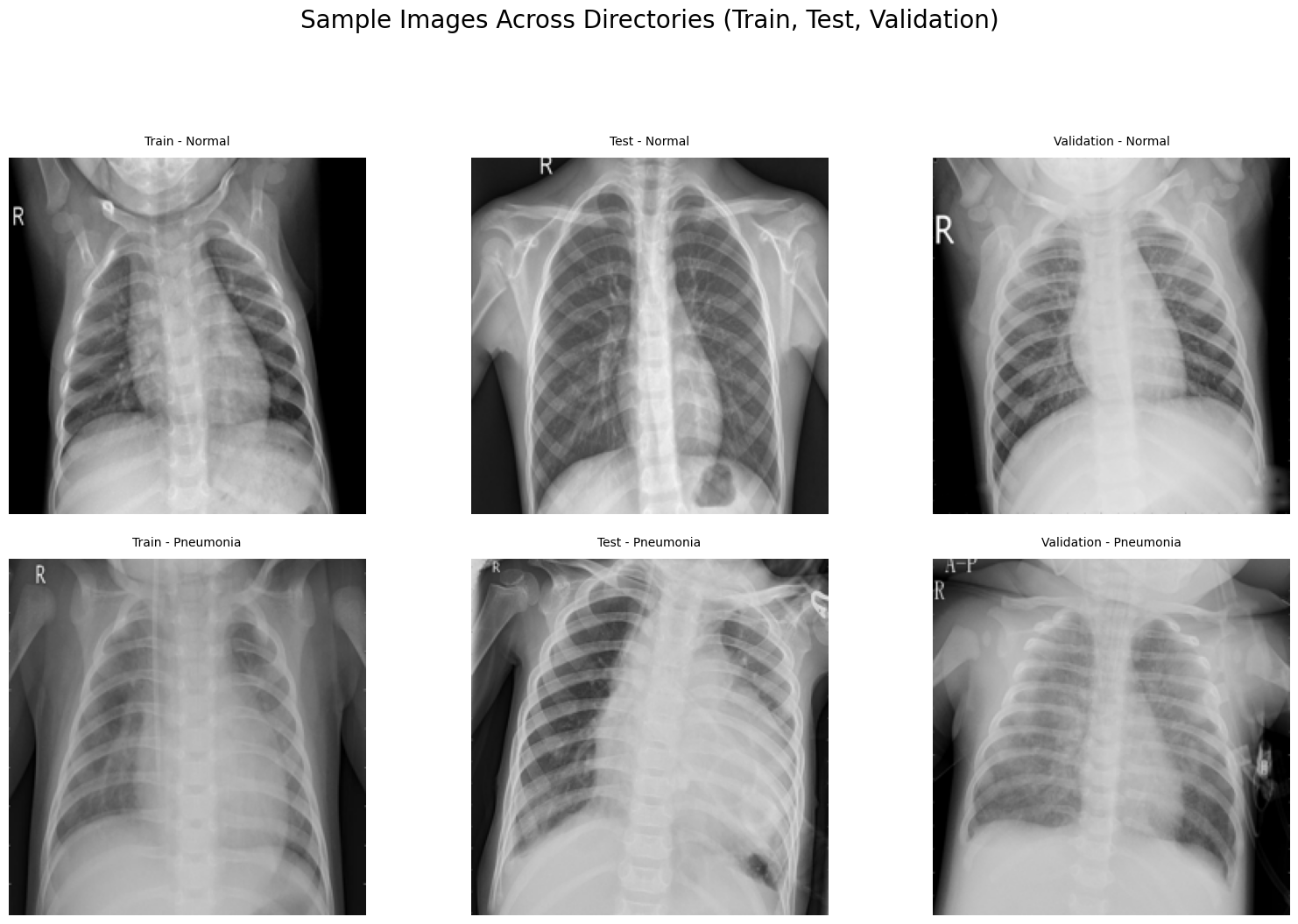
Upon examining the images within the dataset, it becomes evident that, to the untrained eye, distinguishing between normal and pneumonia cases in X-ray images can be quite challenging. However, trained professionals possess the expertise to identify key indicators. This underscores the critical role of modeling and training in enabling the AI to learn these distinguishing features effectively.
To enhance the dataset size, we will first perform data augmentation, incorporating techniques such as random rotations, flips, and zooms. Unlike brain tumor classification, where augmentation can adversely affect model accuracy (Nalepa et al., 2019), the X-ray images in this dataset have been conveniently labeled with an R, which are actually position markers for X-Rays.

The ImageDataGenerator augments and generates the new data for the sets respectively.
ImageDataGenerator Augmented Images Samples (224x224)Using the Augmented dataset, we then proceed to build and train a model to perform classification and segmentation.
Eff-UNet Model
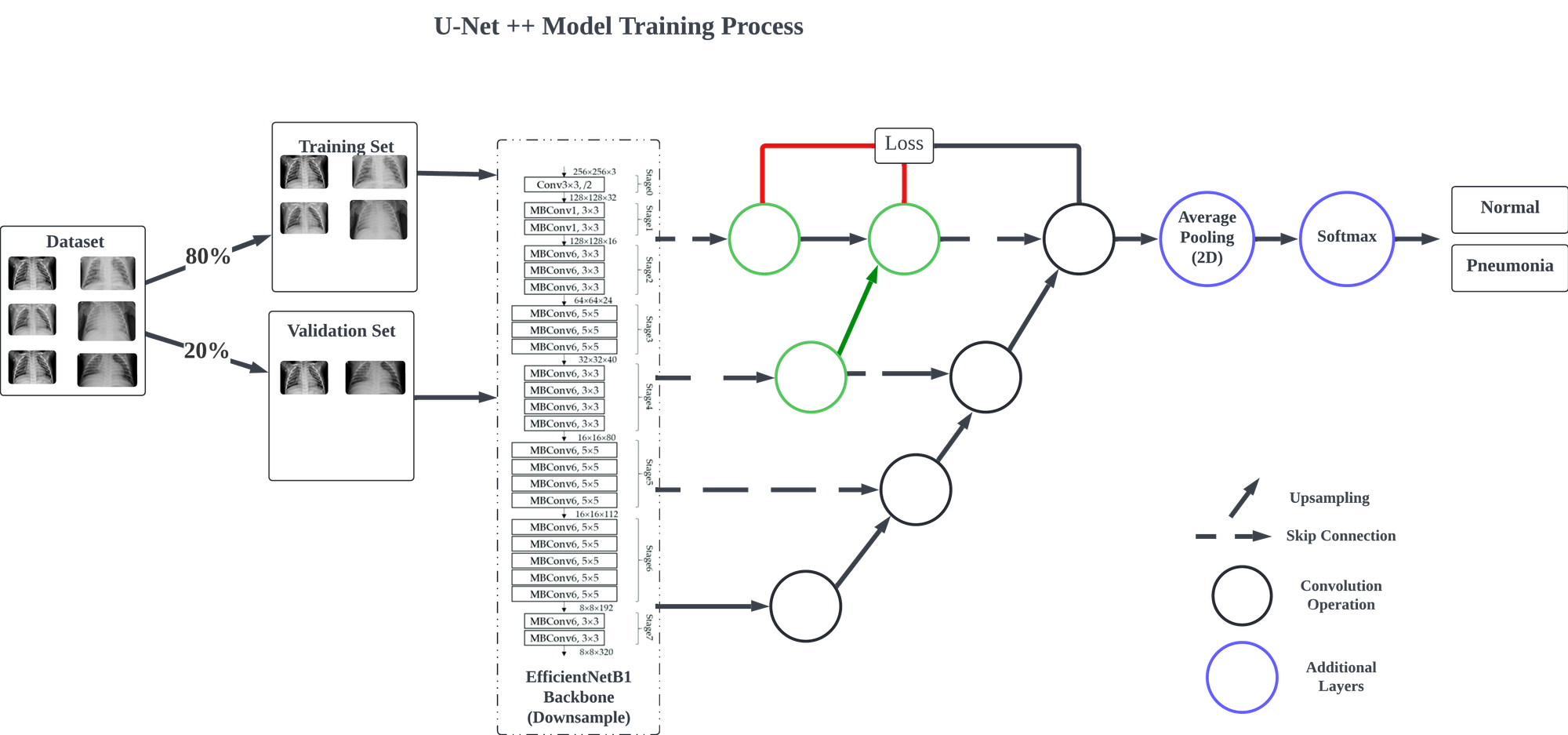
In this model, inspired by methodologies from a previous project, EfficientNetB1 is employed as the primary feature extractor, allowing the model to identify distinguishing features relevant to each classification. The upsampling path is managed by the segmentation_models library, and a Global Average Pooling layer is introduced to streamline the spatial dimensions for classification. Finally, a Dense layer with softmax activation is added to produce the classification output.
The model is compiled using the Adam optimizer with a learning rate of 0.0001 and binary cross-entropy as the loss function. Training and validation are conducted on the designated training and testing datasets, with a final evaluation performed using the validation data generator.
The model was configured to train for 40 epochs, incorporating callbacks such as ReduceLROnPlateau, ModelCheckpoint, and EarlyStopping to optimize the training process. Training concluded after 8 epochs, with the fourth epoch achieving the highest performance.
Analysis
The fourth epoch yielded the best model performance, with a training accuracy of 98.37% and a training loss of 0.1062, indicating effective learning on the training data. However, the validation accuracy dropped to 79.33%, alongside a higher validation loss of 0.4772. This discrepancy suggests potential overfitting, where the model performs well on training data but struggles to generalize effectively to unseen data. Further tuning, such as adjustments in regularization or data augmentation, may be necessary to improve validation performance.
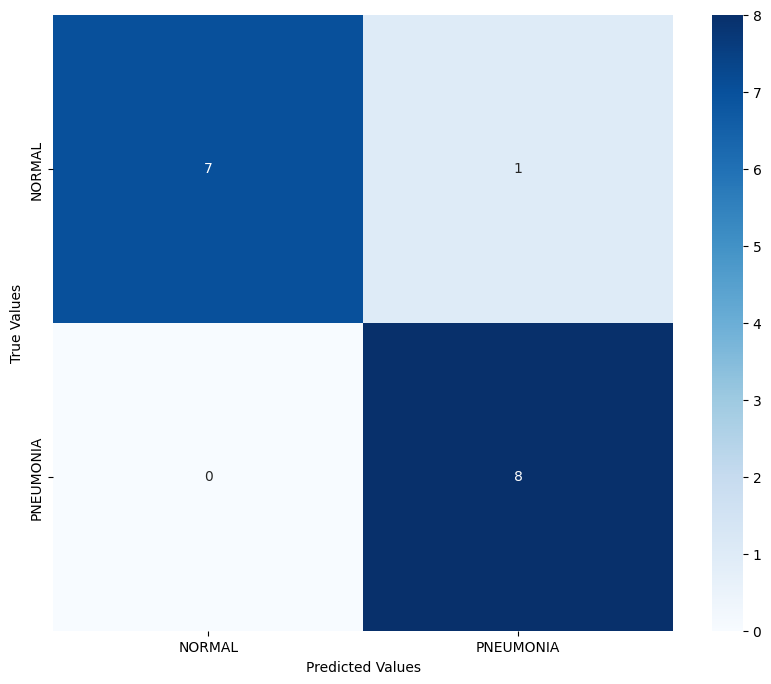
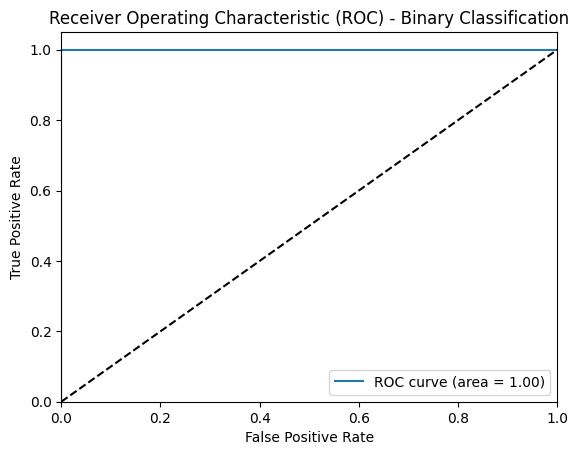
Confusion Matrix & RoC Curve
The confusion matrix reveals the model's strong ability to distinguish between normal and pneumonia cases. For the normal class, the model achieved 7 correct predictions out of 8, with a single false positive where a normal case was incorrectly classified as pneumonia. For the pneumonia class, the model demonstrated perfect recall by correctly identifying all 8 pneumonia cases, with no false negatives. This indicates that the model is particularly effective at detecting pneumonia cases, which is crucial in a medical diagnostic context where missing positive cases (false negatives) could have severe consequences.
The ROC curve further supports this assessment, with an AUC (Area Under Curve) of 1.0, indicating an ideal model with perfect sensitivity and specificity. This high AUC implies that the model maintains consistent performance across varying classification thresholds, demonstrating a robust ability to separate pneumonia cases from normal cases under different conditions.
| Class | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|
| NORMAL | 1.00 | 0.88 | 0.93 | 8 |
| PNEUMONIA | 0.89 | 1.00 | 0.94 | 8 |
| Accuracy | 0.94 | 16 | ||
| Macro Avg | 0.94 | 0.94 | 0.94 | 16 |
| Weighted Avg | 0.94 | 0.94 | 0.94 | 16 |
The classification report provides a quantitative breakdown of the model’s performance. For the normal class, the model achieved a precision of 1.00 and a recall of 0.88, resulting in an F1-score of 0.93. The high precision implies that almost all normal cases predicted by the model were indeed normal, while the slightly lower recall indicates a minor rate of misclassifying normal cases as pneumonia. For the pneumonia class, the model achieved a precision of 0.89 and a recall of 1.00, yielding an F1-score of 0.94. This high recall, coupled with slightly lower precision, indicates that while some false positives occurred, the model was highly sensitive to detecting pneumonia cases. Overall, the model achieved an accuracy of 0.94 across the 16 samples, with both macro and weighted averages for precision, recall, and F1-score standing at 0.94, highlighting the model’s balanced performance across both classes.
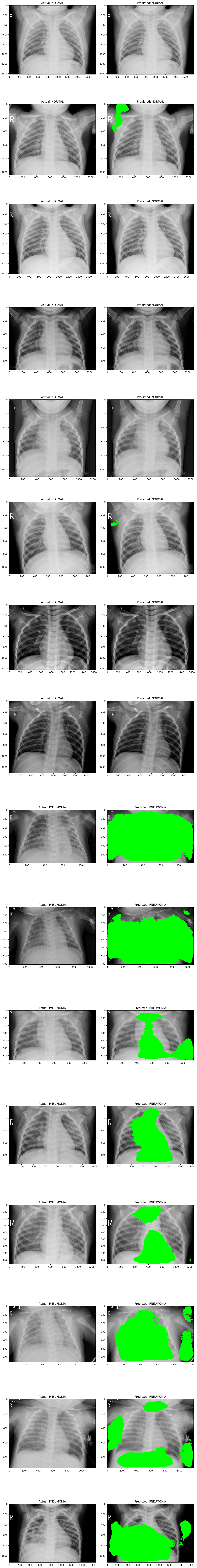
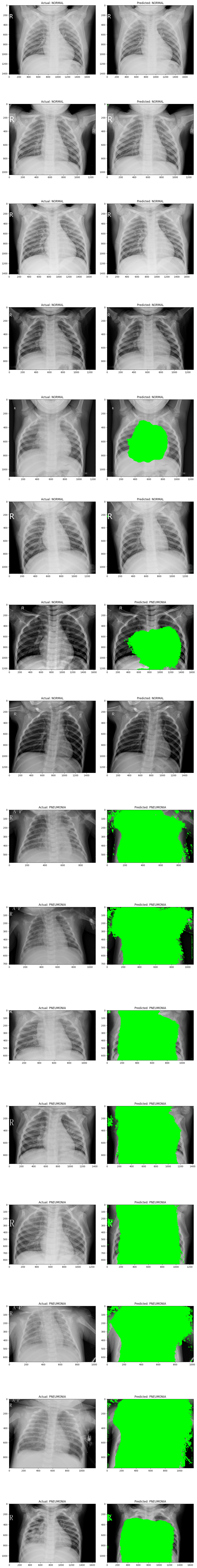
Attempt 1 and 2 respectively
In the segmentation attempt, the model highlights in green the pixels it identified as significant for classification, based on a confidence threshold of 97%. Specifically, any pixel that the model classified with a confidence level of 0.97 or above was marked as part of the relevant features. The segmentation results show that, while the model does identify certain areas as important, there is still an evident gap in its ability to generalize accurately to this task. The model struggled to consistently locate and highlight the areas within the X-ray images that indicate whether the case is normal or pneumonia, suggesting that it hasn't fully learned the distinguishing characteristics.
This limitation implies that further training, possibly with a larger or more diverse dataset, could help the model learn these patterns more effectively. Additionally, fine-tuning the model’s architecture or training parameters may enhance its ability to generalize better to unseen data. However, despite these challenges, the approach shows promise as an assistive tool for healthcare professionals, offering a preliminary classification that radiologists or doctors could verify, potentially accelerating the diagnostic process and improving accuracy.
Conclusion
In this project, we drew on a previous model I developed that effectively identified brain tumors for a specific tumor type, demonstrating the adaptability of the Eff-UNet model architecture for medical imaging.
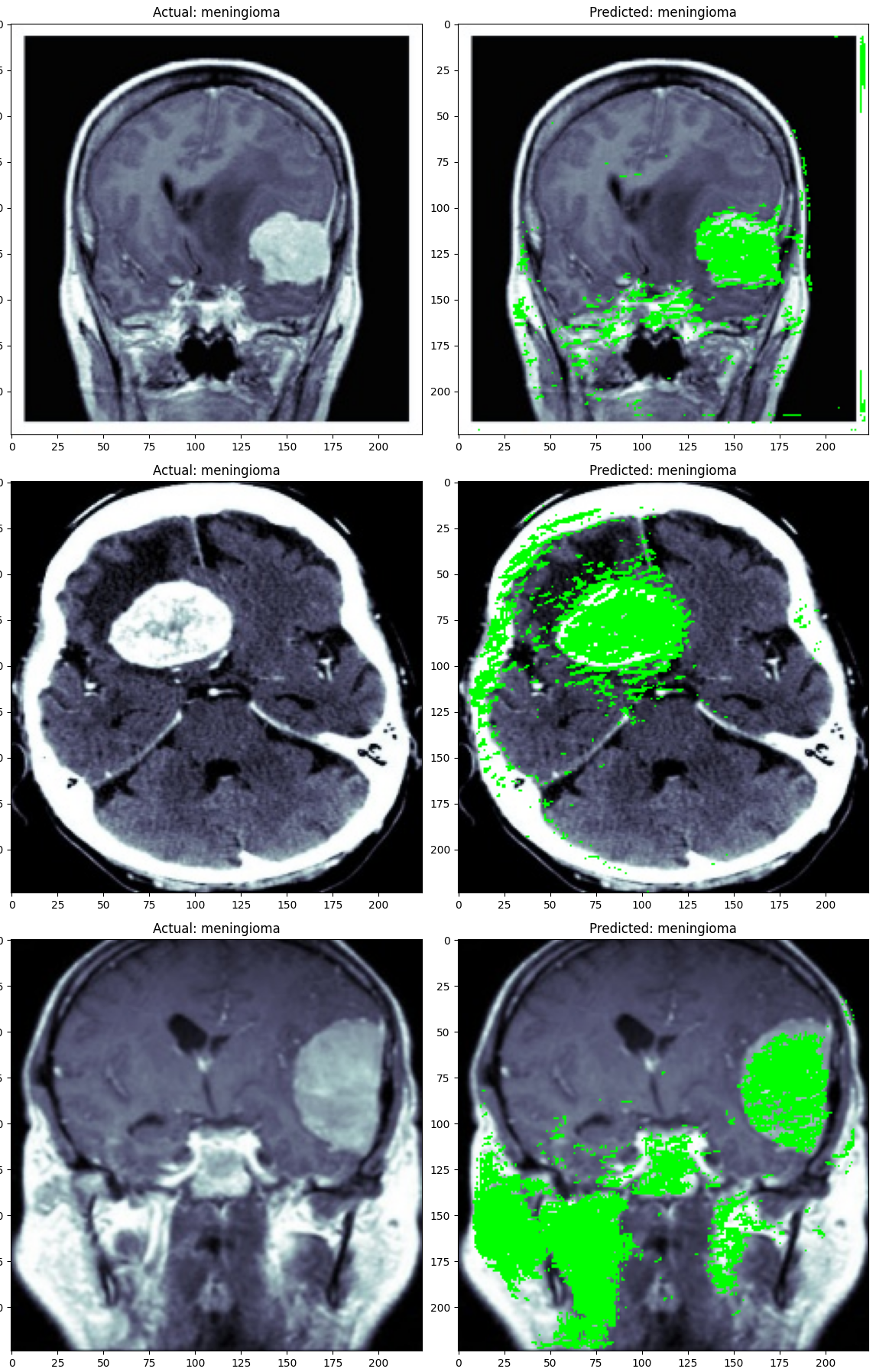
This case reinforces the potential of artificial intelligence and computer vision as valuable aids in healthcare, though they are not yet ready to replace human expertise. With a sufficiently large and well-labeled dataset, the model’s performance would likely have improved significantly, highlighting the importance of data quality and quantity in AI applications for medical diagnostics.
For hospitals and health agencies, a model like Eff-UNet could be trained on extensive databases of medical images, enabling automated anomaly detection and potentially accelerating the diagnostic phase of treatment. In this article, we explored a specific use case for the Eff-UNet model in medical imaging, modifying the model by adding layers for classification purposes, training it on pneumonia detection, and employing segmentation to visualize the regions considered critical for classification. While the results are promising, they highlight the need for further research and model refinement to reach the levels of accuracy and reliability required for real-world clinical application.
References
Abd-Ellah, M. K., Awad, A. I., Khalaf, A. A. M., & Ibraheem, A. M. (2024). Automatic brain-tumor diagnosis using cascaded deep convolutional neural networks with symmetric U-Net and asymmetric residual-blocks. Scientific Reports, 14(1), 9501. https://doi.org/10.1038/s41598-024-59566-7
Baheti, B., Innani, S., Gajre, S., & Talbar, S. (2020a). Eff-UNet: A Novel Architecture for Semantic Segmentation in Unstructured Environment. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 1473–1481. https://doi.org/10.1109/CVPRW50498.2020.00187
Baheti, B., Innani, S., Gajre, S., & Talbar, S. (2020b). Eff-UNet: A Novel Architecture for Semantic Segmentation in Unstructured Environment. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 1473–1481. https://doi.org/10.1109/CVPRW50498.2020.00187
Kermany, D. S., Goldbaum, M., Cai, W., Valentim, C. C. S., Liang, H., Baxter, S. L., McKeown, A., Yang, G., Wu, X., Yan, F., Dong, J., Prasadha, M. K., Pei, J., Ting, M. Y. L., Zhu, J., Li, C., Hewett, S., Dong, J., Ziyar, I., … Zhang, K. (2018). Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell, 172(5), 1122-1131.e9. https://doi.org/10.1016/j.cell.2018.02.010
Lin, S.-Y., & Lin, C.-L. (2024). Brain tumor segmentation using U-Net in conjunction with EfficientNet. PeerJ Computer Science, 10, e1754. https://doi.org/10.7717/peerj-cs.1754
Nalepa, J., Marcinkiewicz, M., & Kawulok, M. (2019). Data Augmentation for Brain-Tumor Segmentation: A Review. Frontiers in Computational Neuroscience, 13. https://doi.org/10.3389/fncom.2019.00083
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation (No. arXiv:1505.04597). arXiv. https://doi.org/10.48550/arXiv.1505.04597
Woon, J. W. (2024). Devoalda/CSC3109_Brain_MRI_Classification [Jupyter Notebook]. https://github.com/Devoalda/CSC3109_Brain_MRI_Classification (Original work published 2024)
Yu, Z. (2021). Pneumonia Detection with U-EfficientNet. 2021 IEEE Sixth International Conference on Data Science in Cyberspace (DSC), 591–594. https://doi.org/10.1109/DSC53577.2021.00093
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2018). UNet++: A Nested U-Net Architecture for Medical Image Segmentation. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support : 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, S, 11045, 3. https://doi.org/10.1007/978-3-030-00889-5_1
Links




Comments ()